Echelon’s ELISAs are centered around biologically interesting lipids and extracellular matrix molecules such as Hyaluronic acid. Lipids are known to be difficult to use as they are hard to solubilize and tend to stick to tubes and vials. Adding to the difficulty, development of ELISA’s generally require a considerable amount of time to maximize the specificity and sensitivity of a binding protein. Detection of the ligand is also complicated by the sample source. Each type of sample brings its own mix of possible cross reactants or interference into the assay. However, this is where Echelon excels, we have 26 years of lipid knowledge and 22 years of ELISA development experience. We utilize this experience with each assay we develop.
An Enzyme-linked immunosorbent assay (ELISA) is a well known and widely used application to detect a specific analyte, commonly a lipid or protein, in a complex mixture or solution. It’s a plate-based assay that relies on an antibody or binding protein to detect this specific target. ELISA’s can be used as a diagnostic tool in medicine or as quality control in the production industry. Echelon offers many ELISA assays that utilize the Direct, Competitive, or Sandwich ELISA assay design. In this blog post, I will give a brief overview of our most common type of ELISA, the Competitive ELISA.
A Competitive ELISA, also known as an inhibition assay, is an ELISA that can measure the concentration of an analyte through its interference in the ELISA assay signal. Competitive ELISAs are great at detecting small analytes such as lipids and can detect these analytes in complex mixtures like plasma, serum, or cell extracts. These assays require less sample processing as they are less sensitive to sample dilution and matrix effects caused from the sample. They tend to have low inter-assay and intra-assay variation as well. Due to this list of reasons, most of our ELISA assays use the Competitive ELISA format.
The assay design for lipid detection
Most of our Competitive ELISA’s are designed for the detection of a specific lipid. In these assays, the lipid is coated onto a multi-well plate. The lipid containing samples are then preincubated with a detection protein that is specific for the lipid you want to detect. The lipid containing sample and detection protein mixture are then transferred to the lipid coated plate. The detection protein and the lipid containing sample will then incubate on the lipid coated plate. During this time, the lipid in the sample will compete with the coated lipid for binding by the detection protein.
Once this incubation is complete, the solution is discarded and the plate is washed. A labeled secondary detection protein is then added. This secondary protein will bind any detection protein bound to the lipid coated plate. The plate is then incubated before the solution is discarded and the plate is washed. Finally, a substrate such as TMB is added to the plate and a colored reaction product is produced. The TMB reaction is stopped and the colored product measured with an absorbance plate reader. This process is outlined in Figure 1 and the quick protocol below.
Quick protocol
- Prepare standards, samples, and detection reagents.
- Add primary detection protein to samples/standards in an incubation plate or tube. Cover plate. Incubate plate or tube 15 – 60 min.
- Transfer solution in the incubation plate to the ligand coated plate. Cover plate. Incubate plate 60 min.
- Wash plate. Add labeled secondary detection protein to ligand coated plate. Cover plate. Incubate plate 30 – 60 min.
- Wash plate. Add substrate and incubate plate 30 min.
- Add Stop solution. Detect signal with absorbance plate reader.
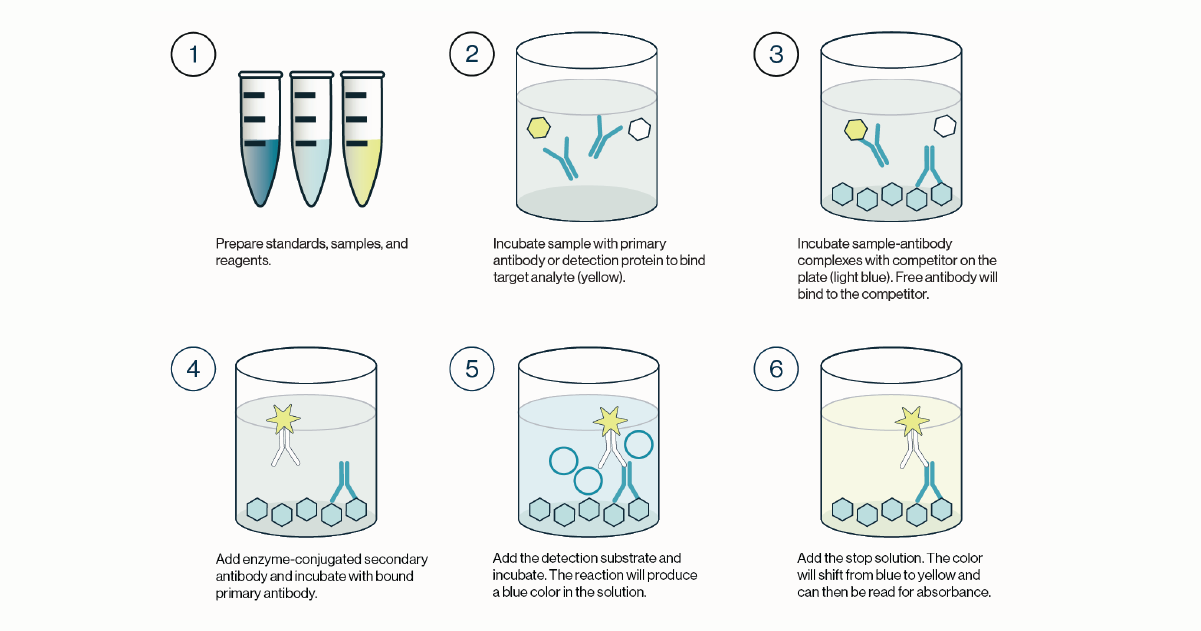
Figure 1: Outline of the procedure for a competitive ELISA. 1) Standards, samples, and reagents are prepared. 2) The sample containing the target analyte is incubated with a primary antibody or detection protein. 3) The resulting complexes from step 2 are transferred to a new plate coated with a competitor that will bind any free detection protein. The remaining solution is discarded and the plate washed. 4) An enzyme conjugated secondary detection protein is added and incubated with the bound primary detection protein. 5) Excess secondary detector is washed and the enzyme substrate is added and incubated. Color development is indicative of detector protein that is bound to the coated competitor. 6) Stop solution is added to quench the enzyme-substrate reaction and the plate is then read for absorbance values.
Analysis of Data
Data from a competitive ELISA can be confusing. The assay signal is opposite to the concentration of the analyte, i.e. inversely proportional. For example, a sample with a low amount of ligand will produce a high assay signal. A sample with a high amount of ligand will produce a low assay signal.
To determine how much ligand is in your sample, the sample optical density (OD) from your plate reader, must be compared to the OD reading of the standard curve. Graphing programs, such as Graphpad, make this analysis easy. If you do not have a graphing program, excel can also do this analysis.
The S1P standard curve below, from the S1P ELISA assay (K-1900), was generated using non-linear regression analysis with GraphPad Software. A semi log [Sigmoidal dose-response (variable slope)] analysis was utilized. For best results, when graphing a competitive ELISA standard curve, constrain the standard curve top & bottom using the 0 (high signal) & Blank (low signal) controls. This ensures accuracy in interpolation of samples that are at the high and low end of the assay window. If you pre-diluted your sample before add to the assay the dilution factor used will need to be applied post interpolation.
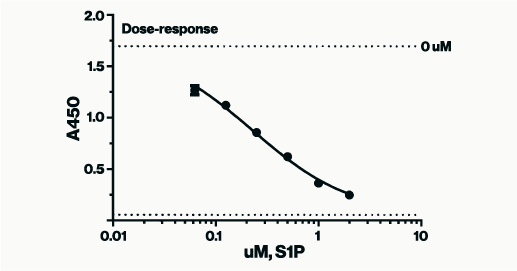
Figure 2: Example of S1P standards in a competitive assay plotted according to concentration vs. absorbance values. As the concentration of the S1P standard is lowered higher absorbance signals are produced. Absorbance values for unknown samples can be interpolated for concentration against a similar curve.
References
1. J R Crowther (2000) The ELISA guidebook. Methods Mol Biol. 149:III-IV, 1-413.