PIP Mass Assays – FAQ
Echelon carries five PIP Mass ELISA kits that can measure the relative levels of 5 different PIPs. Each PIP Mass ELISA uses a protocol for the extraction of phosphoinositides (PIPs) from millions of cells. Extractions are then analyzed using a competitive ELISA without the need for radioactivity.
Download the full PIP Mass Assay FAQ here:
For use with product numbers: K-2500s, K-3300, K-3800, K-4000E, K-4500
Cells
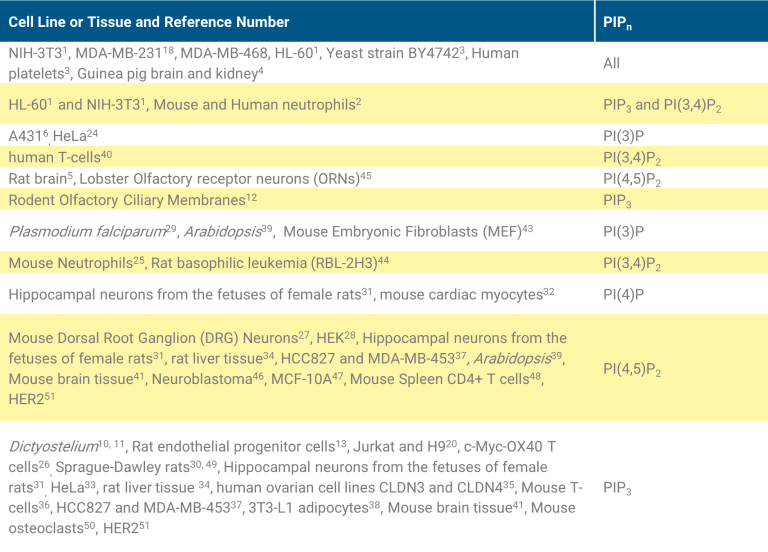
The PIPn extraction protocols have been verified using the cells lines: NIH-3T3, HL-60, MDA-MB-231, and MDA-MB-468. Adherent Cell Lines, NIH-3T3 mouse fibroblasts, MDA-MB-231 human breast cancer, and MDA-MB-468 PTEN deficient human breast cancer were used to validate the assays. The table below lists the published cell lines and tissue samples that have been used in various assays.
Counting Cells
Normalize cells by counting or running a cellular protein assay.
- Adherent cells can be counted before stimulation or before the cell extraction procedure. After counting, return cells to a flask for 1 to 3 hours for re-attachment.
- Non-adherent cells can be counted before stimulation or before the cell extraction procedure. After stimulation, spin cells down, decant the media and add ice cold 0.5 M TCA and proceed with the cell extraction protocol.
- Count or perform protein assay on a separate flask with the same cell dilution as the flasks to be used for cell extraction.
- Counting cells, either by haemocytometer or protein assay, is not recommended after the 0.5 M TCA is added.
- Do not trypsinize cells before adding 0.5 M TCA; it can alter stimulation time and phosphoinositide levels.
- Please refer to the product specific TDS for optimal cell number
Extraction Protocol
The cell extraction protocol has been verified internally at Echelon using the cells lines: NIH-3T3, HL-60, MDA-MB-231, and MDA-MB-468. Please
also see the table under ‘Cells’ for published cell lines and tissues.
- Collect Cells
- For adherent cells, remove medium by gentle aspiration and immediately add ice cold 0.5 M TCA. Incubate cells on ice for 5 minutes. Scrape the cells from flask. Use additional 0.5 M TCA if needed and transfer to a centrifuge tube. Centrifuge at 3,000 RPM (approximately 900-1,000 RCF) for 7 minutes at 4ºC. Discard the supernatant. The remaining steps are performed at room temperature.
- For non-adherent cells, collect cells into centrifuge tube, spin the cells down, decant media, add ice cold 0.5 M TCA and vortex. Incubate cells on ice for 5 minutes. Centrifuge at 3,000 RPM (approximately 900-1,000 RCF) for 7 minutes at 4ºC. Discard the supernatant. The remaining steps are performed at room temperature. Tissue samples should be ground in a mortar and pestle under liquid nitrogen and homogenized before the addition of 0.5 M TCA.
- Wash Pellet: Add 5% TCA/ 1 mM EDTA to the pellet. Vortex for 30 seconds. Centrifuge at 3000 RPM for 5 minutes. Discard the supernatant. Repeat wash one more time.
- It is important to remove as much of the supernatant as possible after final wash. Otherwise, too much water will be present to allow a single phase to form on subsequent extraction steps.
- Extract neutral lipids: Add MeOH: CHCl3 (2:1) and vortex for 10 minutes at room temperature. Centrifuge at 3000 RPM for 5 minutes, discard the supernatant. Repeat neutral lipid extraction one more time. Depending on the number of cells used, a small white pellet should be visible after this step.
- Extract acidic lipids: Add MeOH: CHCl3:12 N HCl (80:40:1) and vortex for 25 minutes at room temperature. Centrifuge at 3,000 RPM for 5 minutes. Transfer supernatant to a new tube. Discard pellet.
- Phase split: To supernatant from step 4, add CHCl3 and then the 0.1 N HCl. Vortex for 30 seconds. Centrifuge at 3,000 RPM for 5 minutes to separate organic and aqueous phases. Disregard any excess cellular debris that may appear between the two layers. Collect the organic (lower) phase, with a positive displacement pipette, into a new vial and dry in a vacuum dryer (45-60 minutes). Dried lipid can be stored at -20ºC for up to 12 months. The dried lipid should not be visible. If there is a visible substance at the end of this step, it is most likely cell debris that was not eliminated in the extraction.
PIPn Extraction Protocol
Volumes are not included in this protocol. Please refer to the specific products TDS for suggested cell number and volumes needed for extraction.
- Collect Cells:
- For adherent cells, remove medium by gentle aspiration and immediately add ice cold 0.5 M TCA. Incubate cells on ice for 5 minutes. Scrape the cells from flask. Use additional 0.5 M TCA if needed and transfer to a centrifuge tube. Centrifuge at 3,000 RPM (approximately 900-1,000 RCF) for 7 minutes at 4ºC. Discard the supernatant. The remaining steps are performed at room temperature.
- For non-adherent cells, collect cells into centrifuge tube, spin the cells down, decant media, add ice cold 0.5 M TCA and vortex. Incubate cells on ice for 5 minutes. Centrifuge at 3,000 RPM (approximately 900-1,000 RCF) for 7 minutes at 4ºC. Discard the supernatant. The remaining steps are performed at room temperature.
- Tissue samples should be ground in a mortar and pestle under liquid nitrogen and homogenized before the addition of 0.5 M TCA.
- Wash Pellet: Add 5% TCA/ 1 mM EDTA to the pellet. Vortex for 30 seconds. Centrifuge at 3000 RPM for 5 minutes. Discard the supernatant. Repeat wash one more time.
- Note: It is important to remove as much of the supernatant as possible after final wash. Otherwise, too much water will be present to allow a single phase to form on subsequent extraction steps.
- Extract neutral lipids: Add MeOH: CHCl3 (2:1) and vortex for 10 minutes at room temperature. Centrifuge at 3000 RPM for 5 minutes, discard the supernatant. Repeat neutral lipid extraction one more time. Depending on the number of cells used, a small white pellet should be visible after this step.
- Extract acidic lipids: Add MeOH: CHCl3:12 N HCl (80:40:1) and vortex for 25 minutes at room temperature. Centrifuge at 3,000 RPM for 5 minutes. Transfer supernatant to a new tube. Discard pellet.
- Phase split: To supernatant from step 4, add CHCl3 and then the 0.1 N HCl. Vortex for 30 seconds. Centrifuge at 3,000 RPM for 5 minutes to separate organic and aqueous phases. Disregard any excess cellular debris that may appear between the two layers. Collect the organic (lower) phase, with a positive displacement pipette, into a new vial and dry in a vacuum dryer (45-60 minutes). Dried lipid can be stored at -20ºC for up to 12 months. The dried lipid should not be visible. If there is a visible substance at the end of this step, it is most likely cell debris that was not eliminated in the extraction.
Extraction Notes
- The amount of cells necessary for PIPn quantification needs to be determined for each cell and PIPn. For a starting point please refer to the
products specific TDS. If larger or smaller amounts of cells are required, over what is specified in the products TDS, the extraction volumes
will need to be proportionately adjusted. - For a PIPn extraction positive control, add 20 pmol diC16 PIPn to step 4, of the PIPn extraction protocol, without cells. Dilute diC 16 PIPn control
lipid in CHCl3: MeOH: H2O (1: 2: 0.8) solution at 0.2 μM. See Table 3 for appropriate diC16 PIPn controls. - If air-displacement pipets are being used to dispense volatile solvents or to recover the lower organic phase, then they should first be well-primed with the organic solvent so the air inside of the pipet becomes saturated with the vapor; otherwise the first few samples will be short-measured.
- Once the extraction solutions are prepared, plastic tips and vials can be used for the remaining steps of the extraction.
- Once step 1 of the extraction protocol is complete, the cell pellet does not need to be on ice.
- If you are running a large number of cell extractions, we suggest staggering the flasks during step 1 and 2 to limit the amount of time the PIPn
is in the TCA solutions. The cell pellet, after the final wash in step 2, can be incubated on ice or at -20ºC for up to 2 hours. Thaw cell pellets
before continuing with the cell extraction protocol or they will not break apart while vortexing. PIPn may not be stable in TCA. - The approximate end volume from the PIPn extraction takes about 1 hour to dry in a speed vacuum dryer. If you do not have a speed vacuum
dryer a nitrogen stream or lyophilizer can be utilized. - The dried lipid should not be visible. If there is a visible substance it is most likely cell debris that was not eliminated during the extraction.
Proceed with detection regardless if a pellet can be seen or not. If the pellet is visible, it may look colorless, white, yellow, brown, or black. Do
not attempt to dissolve visible cell debris in the detection assay buffer. This black to yellow substance should also be avoided when pipetting
the lipid extraction samples into the PIPn Mass ELISA wells. - Due to the variable nature of cells and the difficulty of the PIPn extraction, we recommend running samples in multiples; multiple dilutions,
extractions, and times per condition per data point.
ELISA Detection
ELISA Detection
Once PIPn has been extracted from cellular samples, it is brought up into an aqueous buffer before being incubated with a PIPn detector protein. The PIPn detector /extraction sample are then transferred to the PIPn-coated plate for competitive binding. A peroxidase-linked secondary detection reagent and colorimetric substrate is used to detect the PIPn detector protein that has bound the plate. Since this is a competitive ELISA, the colorimetric signal is inversely proportional to the amount of PIPn extracted from cells. The quantity of PIPn may vary for each cell type, stimulant, and number of cells used.
ELISA Selectivity
The antibodies and binding proteins utilized in each assay have been tested for selectivity of its preferred PIPn over the other synthetic diC 16 PIPn (see Table: Selectivity for the catalog numbers of the lipids tested) and have been found selective for the PIPn they were designed to measure. We developed the kit’s selectivity based on the IC50 values obtained for each PI/PIPn while also considering the relative levels of PIPn found in cells. Please note, that the isolated synthetic diC16 PIPn (shown in Table: Selectivity), may not fully represent the complex mixture of lipids and lipid chain lengths present in a biological extraction.
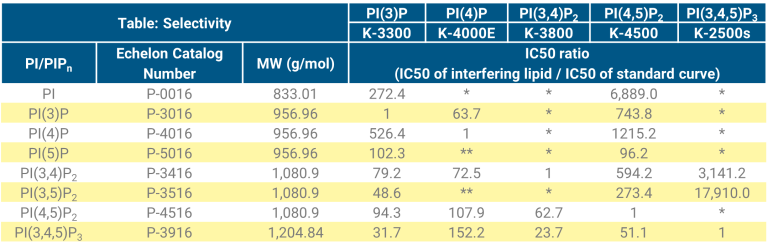
The relative levels of PI/PIPn are dependent upon the cell source, drug treatment, and its disease. For instance, PI(4)P and PI(4,5)P2 constitutes about 2-5% of total PtdIns in mammalian cells53,54, with PI(4)P at ~ 50% the amount of PI(4,5)P2, yet, in yeast and plants cells PI(4)P is found at levels higher then PI(4,5)P255,56. As another example, PI(3)P is found at 5-10% of PI(4)P in mammalian cells but in yeast PI(3)P is found at the same levels of PI(4)P58. PI(3,5)P2 levels in yeast are ~50% of PI(4,5)P259,60 but it is significantly lower in mammalian cells (1% of PI(4,5)P2)61,62. Additionally, PI(3)P3, PI(5)P2, PI(3,5)P2 and PI(3,4,5)P3 are all minor lipids in mammalian cells (1-5% of PI(4,5)P2), that can substantially increase upon activation.
As described above, the cell source and treatment of cells can affect the PIPn profile. Due to this, you may experience interference from a secondary PIPn. This is most often observed with PI(4,5)P2 because of its high level in cells compared to the other PIPn. PI(4,5)P2 cross reactivity is of
greatest concern in the PI(3,4)P2 assay (K-3800). If a sample is highly competitive in an assay and you think a secondary PIPn may be interfering,
the sample should be titrated until it competes in the lower detection range of the assay. The sample should show dilution linearity.
Preparing the PIPn Extraction Sample for Detection in the ELISA Assay
Ensure the dried PIPn extractions tare at room temperature. The room temperature PIPn extractions are then resuspended in assay buffer according to the product specific TDS before being vortexed for 1 min or sonicated for 5-10 min (or both) to solubilize the lipids.
When only vortexing is utilized to dissolve the PIP n extraction sample, lower amounts of PIPn are measured (see Figure 1) and greater day to day variation is observed. The vortexed sample may not show dilution linearity. When the same sample is sonicated, however, we observe greater amounts of measurable PIPn (see Figure 1), consistent day to day variation (inter assay CV = 4.90%) and dilution linearity (2.89% at 1:3 dilution).
It can be difficult, however, to reproduce conditions of sonication; due to variation in the number of vials between batches, temperature of the water bath, and sonicator tuning. The suggested sonication time of 5-10 min was developed using a water bath sonicator. The results observed with your water bath sonicator may be different. It is suggested that your sonicator is tested with extraction samples for day to day variation and time dependent consistency. If you are running other lipid mass assays you may want to consider dissolving your lipids the same manner. How you dissolve the lipid will affect how it goes into solution and can cause inconstancies in your data if it is not held constant.

Other Things to Consider: ELISA Detection
- It is very important for the PIPn Standard and the extraction samples to be at room temperature before they are solubilized. Not doing so will
affect how they behave in the assays. - We have not extensively tested the use of water bath sonication with ice, except to note that less than expected PIPn was measured as
compared to same sample without ice. - Any yellow to black precipitate in the PIPn extraction sample is not PIPn and should be avoided when pipetting the cell samples into the PIPn
Mass ELISA wells. - Cell dilutions higher than six may not work well on the ELISA. We recommend reducing the number of cells used before further diluting the
cell extraction samples. Dilute cell extraction samples as needed.
Assay Quantification
Quantification: PIPn Mass ELISA
To quantify the relative PIPn levels in the PIPn extractions, plot the absorbance (O.D) values obtained vs. PIPn standard to generate a PIPn standard curve. Cellular PIPn levels can then be estimated by comparing the O.D. values of the PIPn extraction samples to the O.D. values on the standard curve.
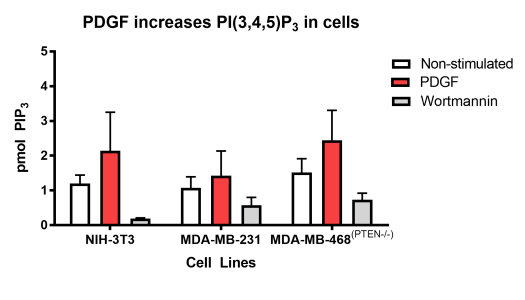
PIP3 Mass ELISA Kit (catalog K-2500s) Example
Figure 2: PI(3,4,5)P3 levels were calculated using Graphpad Prism software and nonlinear regression Sigmoidal Dose Response (variable slope) analysis. Breast cancer cell lines MDA-MB-231 and MDA-MB-468 (PTEN-/-) were extracted from 12-well plates and run on the PIP3 Mass ELISA Kit (K-2500s). The cells were serum starved for 5 hours before they were stimulated with 10 ng/mL PDGF for 5 min or 300 nM Wortmannin for 3 hours respectively.
Sample normalization with PI(4,5)P2
PI(4,5)P2 is the most abundant PIPn in mammalian cells and therefore can be used to normalize samples by using the PI(4,5)P2 measurement as a indicator of total phosphoinositide. This method has been used by Costa et al. for the measurement of PI(3,4,5)P3. This group justified the use of this PIPn over the others due to its high abundance and PI3K’s lack of influence over its levels. To use this control each extraction sample will need to be split into two vials at the end of the extraction, before the sample is dried. For example: if 1.5 mL of the lower (organic) layer is collected then 1.45 mL is transferred to one vial for PI(3,4,5)P3 detection in the PIP3 Mass ELISA (K-2500s) and 0.05 mL is transferred to another vial for PI(4,5)P2 detection in the PI(4,5)P2 mass ELISA (K-4500). These are both dried and then brought up according to the protocol of the specific PIPn Mass ELISA kit being used. Once the ELISA’s are run and the results quantified the pmol values are then entered into this formula:
PI(3,4,5)P3/PI(4,5)P2 = % total PI(4,5)P2