Lipid beads are tools designed for studying lipid-protein interactions. Initially developed with eight phosphatidylinositol phosphate coatings, they now feature a total of 28 lipid surface coatings. These beads were primarily intended for protein pull-down experiments to validate results from lipid-protein overlay assays (Lipid Strip). However, recent advancements in lipidomic and proteomic technologies have enhanced the ability to investigate protein-lipid interactions in cell lysates, nuclear extracts, and other cellular components. Lipid beads can used in this exploratory pipeline.
'Fishing' for lipid-protein interactions
The use of immunoprecipitation, affinity captured, or immobilized proteins to identify potential binding partners via mass spectrometry (mass spec) and proteomics is a well-established methodology. A generic version of this procedure will use a purified protein to capture putative binding partners from a cell lysate to identify novel interactions. However, this strategy has only recently been applied to lipids as increasing numbers of non-lipid biologists recognize their importance beyond membrane structure.
Historically, most lipid-binding proteins have been identified by a 'bottom-up' approach, wherein their potential association with lipids is inferred by cell imaging techniques and then later confirmed by a combination of bioinformatics, biochemistry, and molecular biology. The converse, 'top-down' or 'fishing' approach described above has been more difficult to implement for lipids as lipid-protein interactions may be lost upon cell lysis or during the steps required to purify and isolate target proteins.
These concerns can be circumvented by immobilizing a single lipid class or species to a solid support or resin, which is the approach taken with lipid beads. In this manner, the beads can be used in a mode similar to an antibody or protein conjugated to a solid support to capture proteins from complex lysate solutions.
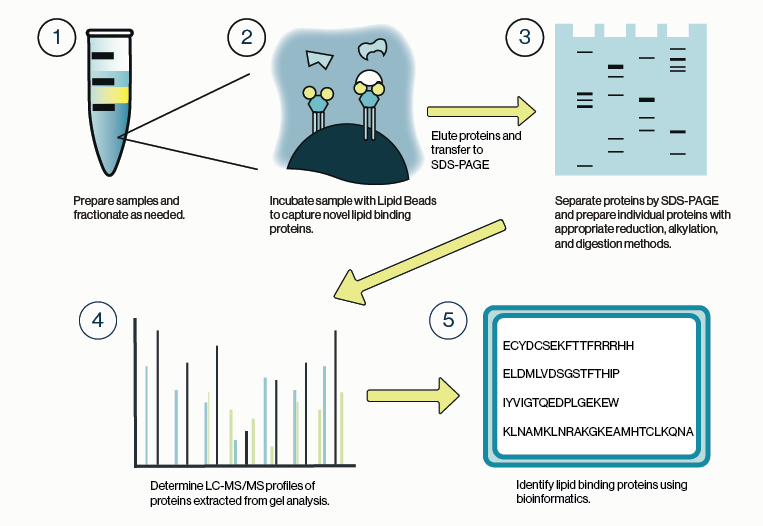
Figure 1: Workflow for identification of protein-lipid binding partners from cell lysate using Echelon lipid coated beads and LC/MS analysis.
Workflow for Lipid Beads and Mass Spec
The general process for capturing lipid-binding proteins is highly similar to that for protein-protein interactions, although there are some special considerations. Chief among these is the stringency of the buffers used for harvesting and lysis of cells or tissue. Ionic detergents such as SDS are commonly used to solubilize lipid and membrane components of the cell but, should be avoided here as they may obscure protein interactions. Non-ionic detergents such as Igepal or Triton are preferable in this case as they are less likely to interfere with lipid binding. Similarly, it is advisable to maintain the salt concentration of any lysis buffer close to physiologic levels.
Once the samples are sufficiently lysed and fractionated, the lipid beads can be applied to the samples. Care should be taken to avoid overloading the beads; limit the protein concentration to 1 mg/mL or less. Additionally, shorter incubation times of 1-3 hours are recommended. These two points can help to decrease non-specific binding to the beads and will limit identification of protein artifacts during mass spec analysis.
It is also recommended that one or more control conditions be run in parallel with the target lipid bead. The simplest control would be to run a non-coated bead or a bead coated with a different lipid species. An alternative would be to pre-incubate the sample with a molar excess of the target lipid to fully compete binding to the lipid bead. These types of controls provide a background mass spec profile that can be subtracted during the bioinformatic analysis.
Confirmation and Follow Up Experiments
In recent years there has been a push from the mass spec community to treat results from some types of 'omics' experiments as independent and not requiring external validation. While this view is well founded, it is never a bad idea to have multiple methods or results pointing to the same conclusion.
Several types of experiments can be used to confirm lipid-protein interactions. These include, but are not limited to, sedimentation assays, liposome flotation, and lipid-protein overlays. We have covered best practices for overlay assays on this blog. Other protocols for liposome preparation or use of lipid beads can be found here. Happy fishing!
References
- Kattan, R et al. (2022) Interactome analysis of human phospholipase D and phosphatidic acid-associated protein network. Molecular & Cellular Proteomics 21 (2)
- Durrant, T et al. (2019) Identification of PtdIns(3,4)P2 effectors in human platelets using quantitative proteomics. BBA – Molecular and Cell Biology of Lipids, 1865.
- Zhu, Z et al. (2019) Ceramide regulates interaction of Hsd17b4 with Pex5 and function of peroxisomes. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids.
0.2
/ 0.3
Related Articles
Stay informed with our informative blog posts.
0.3
/ 0.3
Get in Touch
If you have any questions or would like to learn more about our services, feel free to reach out to us. We’re here to help!
Biosciences