Phospholipid Enzymes in Cancer
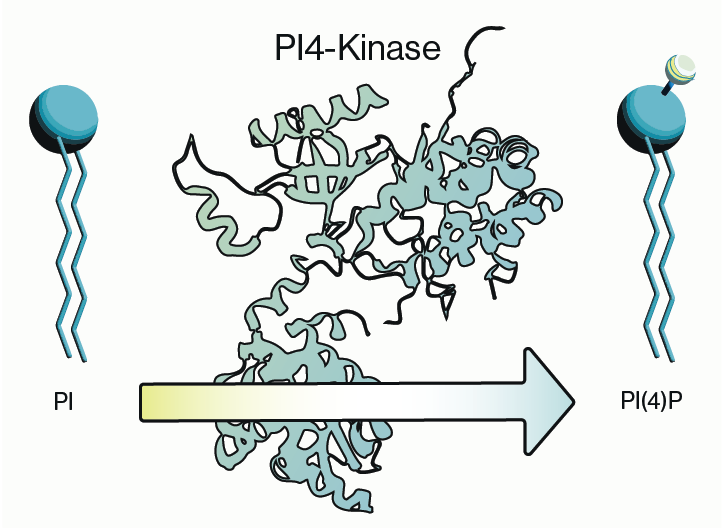
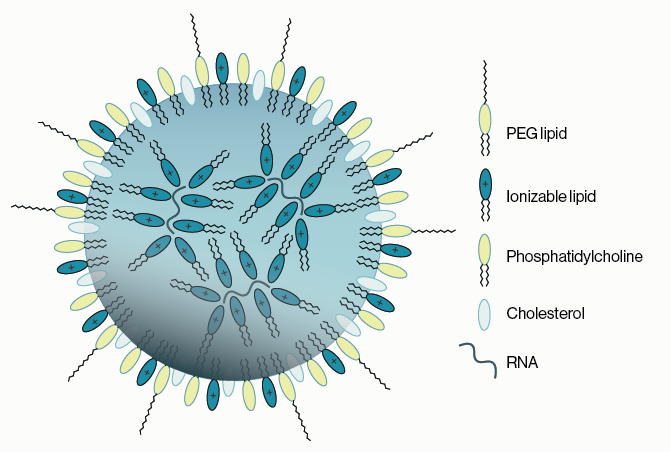
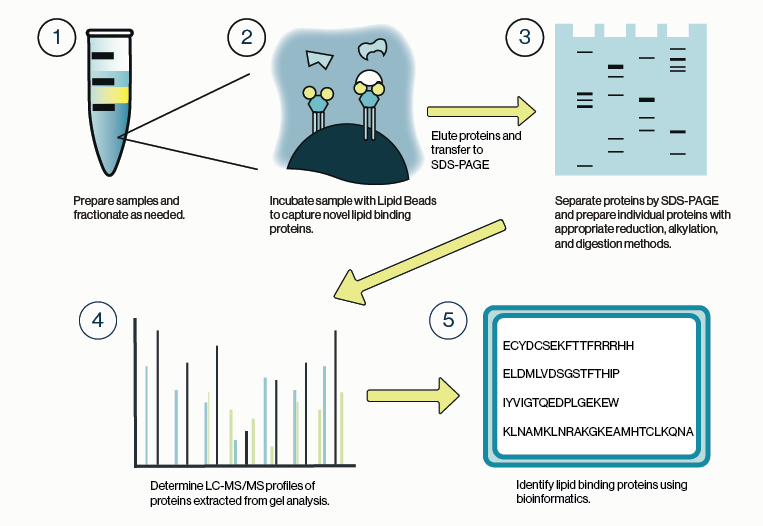
Phospholipids were once thought to be pedestrian structural components of cell membranes. Researchers have since discovered that specialized lipids (including phosphorylated inositol lipids, phosphoinositides, PIPs) are essential for proper cell division, growth, and migration. These properties make phosphoinositides prime candidates for cell transformation involved in cancer. Indeed the enzymes involved in making PIPs, PIP kinases, and particularly PI3K were found to be among the most commonly mutated oncogenes in solid tumors. Further, a lipid phosphatase gene PTEN, which breaks down PIPs, is one of the most frequently inactivated tumor suppressor genes. Together these discoveries place the PIP kinase and phosphatase enzymes at the center stage of cancer drug discovery.
Class 1 PI3-Kinases
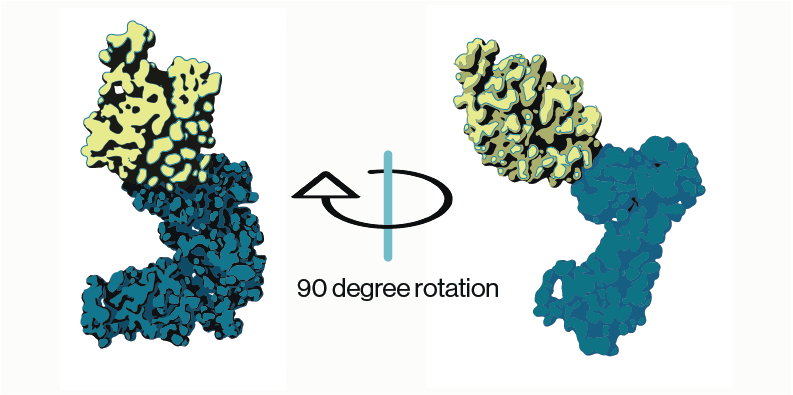
The Class 1 PI3-kinase family consists of catalytic and regulatory subunits each with 5 domains and coded by separate genes. Once formed, the two subunits function as tightly-associated heterodimers inside of cells to transduce extracellular pro-growth signals in the cytosol and eventually to the nucleus. Humans have three isoforms of Class 1A enzymes termed alpha, beta, and delta (the gene names are PIK3CA, PIK3CB, and PIK3CD). Humans have one Class 1B isoform, called gamma (PIK3CG). The p110 catalytic subunits of Class 1A and 1B PI3-Kinases partner with specific regulatory subunits, p85a/p85b/p55 for the 1A enzymes and predominantly p101 for the 1B enzyme. No matter the names and partners, these kinases are primarily grouped because of their catalytic function—they are lipid kinases, and each one catalyzes the addition of a third phosphate to the inositol phospholipid head group of PIP2 to produce PIP3 (Figure 1).
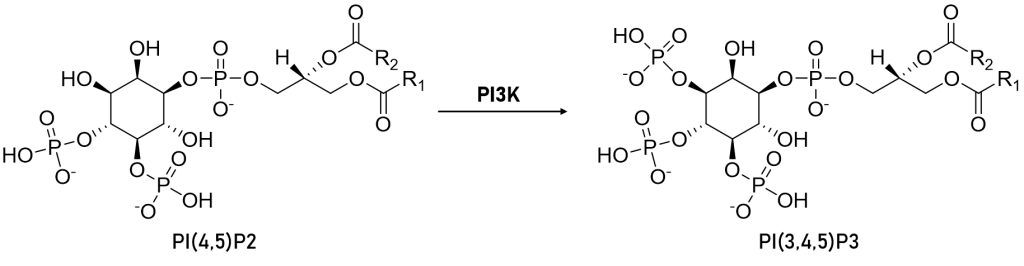
Figure 1: Simple schematic of the chemical structures of the site of action of PI3K on PI(4,5)P2 - phosphate is added to the 3-position on the head group thus generating PI(3,4,5)P3.
Oncogenic PIK3CA Mutants
The p110-alpha catalytic subunit was first identified as an oncogene in 2004 by Yardena Samuels and colleagues in the labs of Bert Vogelstein and Victor Velculescu. Under normal physiologic conditions, PI3K is activated in response to a variety of growth factors and extracellular stimuli. As shown above in Figure 1, following activation, PI3K catalyzes the conversion of PIP2 to PIP3. PIP3 can then act as a second messenger to recruit other kinases such as PDK1 and AKT to activate cell survival pathways. It is now understood that mutations within the kinase domain of p110 lead to constitutive activity of PI3K and these so called 'activating mutations' are a leading cause of tumorigenesis.
The initial discovery of PIK3CA mutations in colon cancer led to the examination of PIK3CA mutations in additional cancer types with the Cancer Genome Atlas identifying PIK3CA (and PTEN) among the genes most frequently developing somatic point mutations in solid tumors. Interestingly activating mutations appear to cluster in two “hotspot” regions in the helical and kinase domains (Figure 2). Mutations in the helical domain drives molecular disinhibtion of PI3K by decreasing the regulatory effect of p85. The other cluster of mutations in the kinase domain promotes association of PI3K with membranes. Both types of mutant increase activity of PI3K and they have been observed to act synergistically when both are present. For these reasons, PI3K activity and inhibition of PI3K has been a longstanding target for cancer therapy.
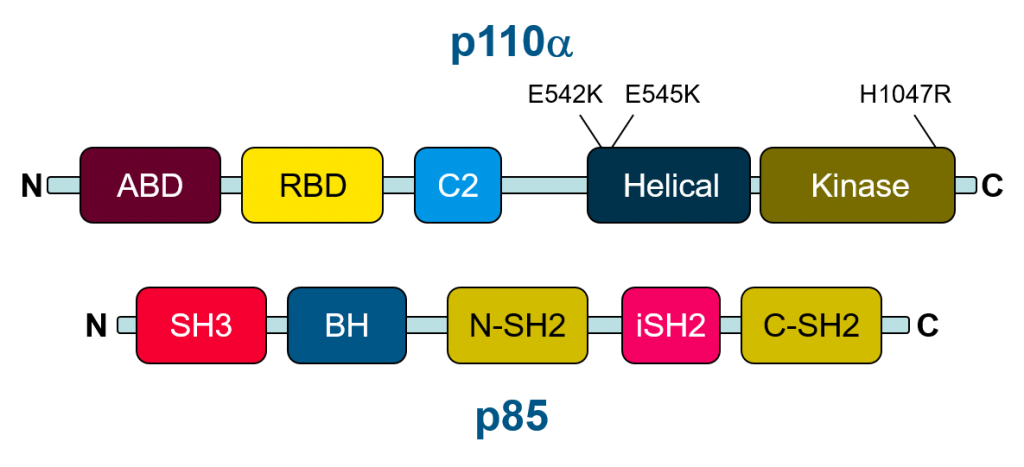
Figure 2: schematic of the domains found within the major PI3K subunits p110 alpha and P85. Common 'hotspot' mutations in the PIK3CA gene are found in the helical and kinase domain of the p110 subunit.
Therapeutic Outlook
Inhibtion of PI3K is an ongoing area of clinical development. Several dozen PI3K inhibtors in different subclasses are currently in clinical trials. An even longer list of prospective trials are currently recruiting patients. Very few of these small molecule drugs have received approval for use as therapies. Copanlisib and duvelisib are two currently approved PI3K inhibitors for treatment of follicular lymphoma and chronic lymphocytic leukemia, respectively. In May 2019 Novartis earned FDA approval for the first breast cancer drug (alpelisib, Piqray) to target tumors harboring PIK3CA mutations. Approval was also given for its companion diagnostic test allowing oncologists to develop treatment plans based on the genomic profile of individual patient’s cancer. Notably, these compounds act on different isomers of PI3K with alpelisib being specific for PI3K alpha while copanlisib and duvelisib act on alpha-beta and gamma-delta.
While it has been exciting to see how far the science of phospholipids and cutting edge cancer treatment has advanced, there remain unmet challenges with the development of PI3K based treatments. This is underscored by the relative specificity for various PI3K isoforms as discussed above. It is also increasingly clear from the trial data that PI3K inhibitors may be best used in combination therapy approaches. Therefore, additional work is required to understand specific mechanisms of PI3K inhibition, isoform-selectivity, and how tumor response is achieved.
References
1. Yang J, Nie J, Ma X, Wei Y, Peng Y, Wei X. (2019) Targeting PI3K in cancer: mechanisms and advances in clinical trials. Molecular Cancer. 18(1):26.
2. Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. (2004) High Frequency of Mutations of the PIK3CA Gene in Human Cancers. Science. 304(5670):554
3. André F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. (2019) Alpelisib for PIK3CA-Mutated, Hormone Receptor–Positive Advanced Breast Cancer. New England Journal of Medicine. 380(20):1929-40.
0.2
/ 0.3
Related Articles
Stay informed with our informative blog posts.
0.3
/ 0.3
Get in Touch
If you have any questions or would like to learn more about our services, feel free to reach out to us. We’re here to help!
Biosciences