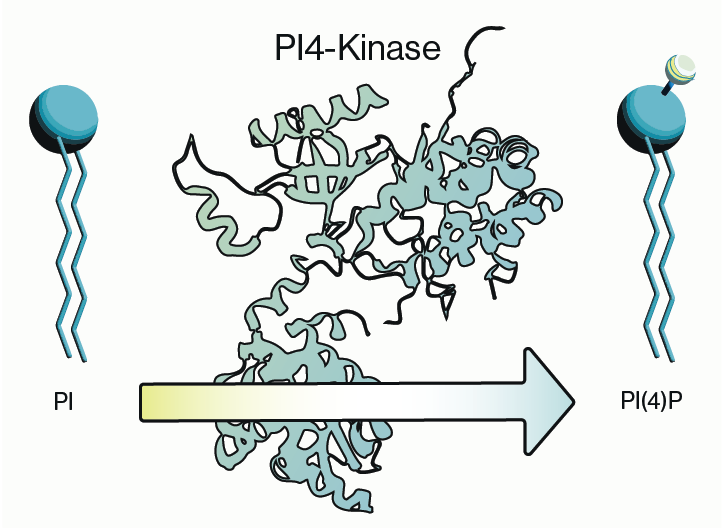
PIKfyve, also known as PIP5K3, is a lipid kinase that synthesizes two rare phosphoinositides, PI(5)P and PI(3,5)P2 (Figure 1). The synthesis of these two phospholipids coordinates specific signaling cascades within the endosome-lysosome-autophagy axis in the cell and dysfunction within this system is observed in several diseases making PIKfyve an attractive therapeutic candidate. PIKfyve has been implicated in several types of cancer and there is a growing body of evidence that its activity is involved in neurodegenerative disorders. More recent interest in PIKfyve has surged due to some evidence showing efficacy of a PIKfyve inhibitor, Apilimod, against SARS-CoV-2 entry into cells. Here we will briefly summarize PIKfyve function and how inhibiting its activity may be utilized for treatment of some diseases.

Figure 1: Synthesis pathways involving PIKfyve for PI(5)P and PI(3,5)P2.
PIKfyve Function
PIKfyve function was initially characterized in yeast where knockdown of its homologue caused a notable phenotype in the form of large vacuoles. Subsequent studies found that this was at least partially due to disruption of ion homeostasis within the lysosome. This observation was later directly tied to synthesis of PI(3,5)P2 which was shown to bind to lysosomal cation channels. Additional evidence found that the enlarged vacuole/lysosome phenotype was also due to membrane trafficking defects between endosomes and lysosomes which suggested PIKfyve function extended throughout the endosomal-lysosomal system. Hence synthesis of PI(3,5)P2 via PIKfyve is critical for activation of membrane channels within the lysosome that regulate fission and formation.
PIKfyve’s role in the endosome was further clarified by the finding that it formed a complex with two other proteins, ArPIKfyve (Vac14) and Sac3 (Fig4), that bound PI(3)P on Rab5-associated endosome membranes. PIKfyve was later shown to colocalize on these membranes with elements of the Retromer complex which is critical for endosomal trafficking. Interestingly, Sac3/Fig4 is a phosphatase that converts PI(3,5)P2 back to PI(3)P implying that the synthesis of PI(3,5)P2 is transient and very tightly regulated. Adding to the complexity is the dependence on PI(3)P for membrane association. PI(3)P is produced by the Class III PI3-Kinase Vps34 and loss of its activity has downstream effects on PIKfyve activation.
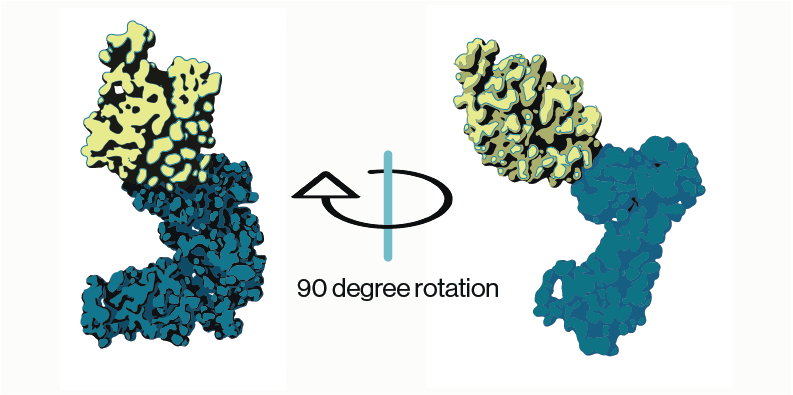
Additional insight into the PIKfyve regulatory complex was gained from structural work on the three proteins. CryoEM reconstructions of of the PIKfyve-Vac14-Fig4 complex showed that Vac14 acted as a pentameric scaffolding protein to support association of PIKfyve and Fig4 on membranes. Curiously, while the CCT domain of PIKfyve (Figure 2) could be directly associated with Vac14, these structures oriented PIKfyve’s kinase domain away from the membrane suggesting that other conformations must be possible. This is supported by additional biochemical work from multiple studies that show PIKfyve and Fig4 reciprocally regulate their activity.

Figure 2: PIKfyve structure (adapted from Lees and Li et al. 2020; PDB entry 7K2V). Model shows cryo-EM densities for predicted PIKfyve’s CCT (yellow) and kinase (blue) domains.
Disease Association & PIKfyve Inhibitors
PIKfyve has been associated with several diseases including some types of cancer and various forms of neurodegeneration. It should be noted though that some of this dysfunction is attributable to mutations in proteins that associate with PIKfyve. For example, a type of Charcot-Marie-Tooth (CMT) syndrome is caused by mutations in Fig4. However, as stated above, PIKfyve and Fig4 have activity towards each other so mutant Fig4 could lead to excess activity from PIKfyve. From this perspective, PIKfyve inhibition could be therapeutic.
Currently there is a great deal of interest in PIKfyve inhibitors, although there are only two widely used molecules. One of these, apilimod, was first identified several years ago as an inhibitor of cytokine release via toll-like receptors leading to clinical trials for various inflammatory disorders. While it failed to show efficacy in these trials, the drug was well tolerated and once PIKfyve was identified as the molecular target, apilimod became more broadly used in basic science research.
More recent work has found that inhibition of PIKfyve via apilimod blocks host cell entry of viruses such as Ebola and SARS-CoV-2 and application of another inhibitor, YM201636, improves cell survival in models of amyotrophic lateral sclerosis. Subsequent studies found that YM201636 PIKfyve inhibition altered the development of tau aggregates in the lysosome, which are associated with some types of neurodegeneration. Additionally, emerging work has shown that inhibition via anti-sense oligonucleotides (ASO) targeting PIKfyve initiates a novel mechanism in the cell where by protein aggregates are cleared by exocytosis through the multivesicular body-exosome pathway.
Whether these small molecule inhibitors can be translated to the clinic remains to be seen, but it is increasingly clear that pharmacological manipulation or blockade of PIKfyve activity has great therapeutic potential. Furthermore, given the growing body of work regarding PIKfyve complexes, there are additional proteins that may serve as targets for similar therapeutic outcomes.
References
- Lees J, Li P, Kumar N, Weisman L, Reinisch K (2020) Insights into Lysosomal PI(3,5)P2 Homeostasis from a Structural-Biochemical Analysis of the PIKfyve Lipid Kinase Complex. Molecular Cell 80(4):736-743
- Rivera-Rios P & Weisman L (2022) Roles of PIKfyve in multiple cellular pathways. Curr Opin Cell Bio. 76
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. (2004) High Frequency of Mutations of the PIK3CA Gene in Human Cancers. Science. 304(5670):554
- Ikonomov O, Sbrissa D, Shisheva A (2019) Small molecule PIKfyve inhibitors as cancer therapeutics: Translational promises and limitations. Tox App Pharma. 383:114771
- André F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. (2019) Alpelisib for PIK3CA-Mutated, Hormone Receptor–Positive Advanced Breast Cancer. New England Journal of Medicine. 380(20):1929-40.
0.2
/ 0.3
Related Articles
Stay informed with our informative blog posts.
0.3
/ 0.3
Get in Touch
If you have any questions or would like to learn more about our services, feel free to reach out to us. We’re here to help!
Biosciences